Operations that support range-partitioning in Modin#
The following operations change their behavior once cfg.RangePartitioning variable is set to True.
Go through the list find out when it could be beneficial to engage range-partitioning for a certain method.
GroupBy#
Note
When grouping on multiple columns using range-partitioning implementation, the result
may not be sorted even if groupby(sort=True, ...) was passed: modin-project/modin#6875.
Range-partitioning groupby implementation is automatically engaged for groupby.apply(), groupby.transform(),
groupby.rolling(). For groupby aggregations from this list, MapReduce implementation is used by default.
MapReduce tends to show better performance for groupby with low-cardinality. If the cardinality of your columns
to group is expected to be high, it’s recommended to engage range-partitioning implementation.
Merge#
Note
Range-partitioning approach is implemented only for “left” and “inner” merge and only when merging on a single column using on argument.
Range-partitioning merge replaces broadcast merge. It is recommended to use range-partitioning implementation if the right dataframe in merge is as big as the left dataframe. In this case, range-partitioning implementation works faster and consumes less RAM.
Under the spoiler you can find performance comparison of range-partitioning and broadcast merge in different scenarios:
Performance measurements for merge
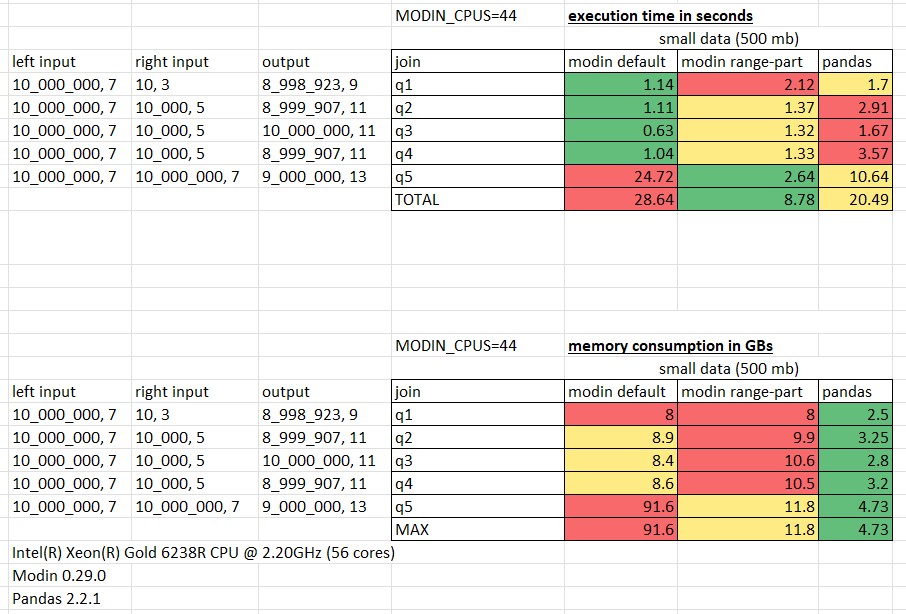
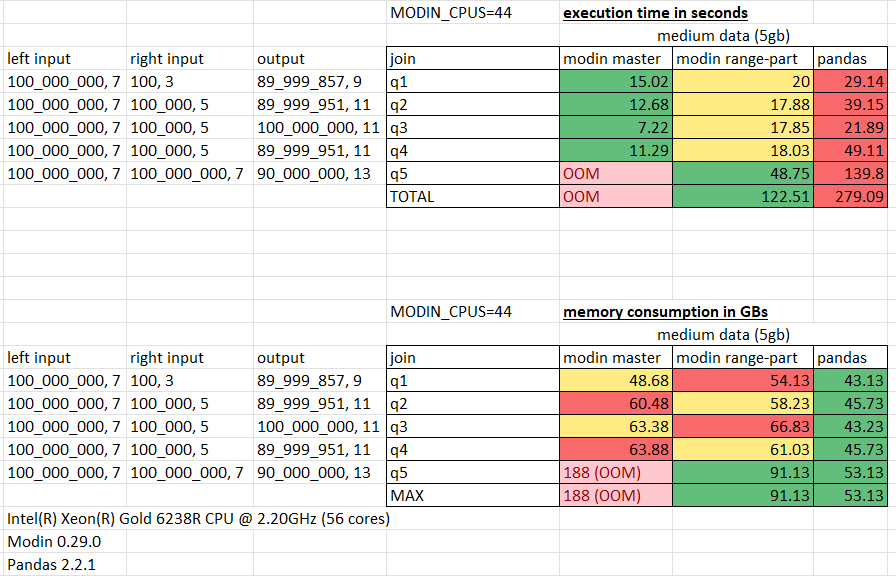
The performance was measured on h2o join queries using Intel(R) Xeon(R) Gold 6238R CPU @ 2.20GHz (56 cores),
with the number of cores allocated for Modin limited by 44 (MODIN_CPUS=44).
Measurements for small 500mb data:
Measurements for medium 5gb data:
.unique() and .drop_duplicates()#
Note
When range-partitioning is enabled, both .unique() and .drop_duplicates() will
yield results that are sorted along rows. If range-partitioning is disabled,
the original order will be maintained.
Range-partitioning implementation of .unique() / .drop_duplicates() works best when the input data size is big (more than
5_000_000 rows) and when the output size is also expected to be big (no more than 80% values are duplicates).
Under the spoiler you can find performance comparisons in different scenarios:
Performance measurements for ``.unique()``
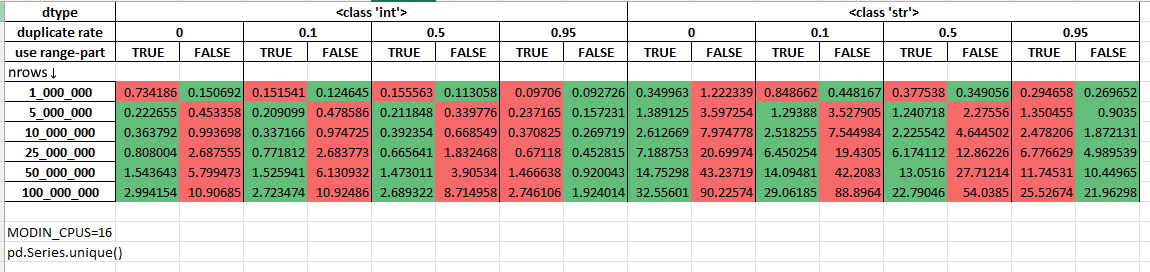
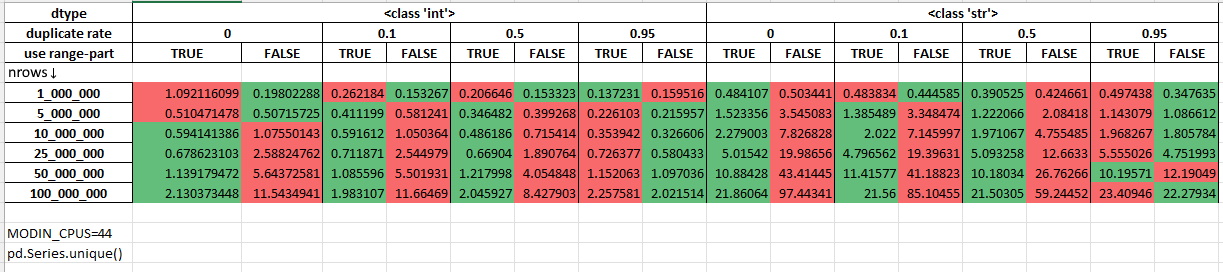
The performance was measured on randomly generated data using Intel(R) Xeon(R) Gold 6238R CPU @ 2.20GHz (56 cores). The duplicate rate shows the procentage of duplicated rows in the dataset. You can learn more about this micro-benchmark by reading its source code:
Micro-benchmark's source code
import modin.pandas as pd
import numpy as np
import modin.config as cfg
from modin.utils import execute
from timeit import default_timer as timer
import pandas
cfg.CpuCount.put(16)
def get_data(nrows, dtype):
if dtype == int:
return np.arange(nrows)
elif dtype == float:
return np.arange(nrows).astype(float)
elif dtype == str:
return np.array([f"value{i}" for i in range(nrows)])
else:
raise NotImplementedError(dtype)
pd.DataFrame(np.arange(cfg.NPartitions.get() * cfg.MinPartitionSize.get())).to_numpy()
nrows = [1_000_000, 5_000_000, 10_000_000, 25_000_000, 50_000_000, 100_000_000]
duplicate_rate = [0, 0.1, 0.5, 0.95]
dtypes = [int, str]
use_range_part = [True, False]
columns = pandas.MultiIndex.from_product([dtypes, duplicate_rate, use_range_part], names=["dtype", "duplicate rate", "use range-part"])
result = pandas.DataFrame(index=nrows, columns=columns)
i = 0
total_its = len(nrows) * len(duplicate_rate) * len(dtypes) * len(use_range_part)
for dt in dtypes:
for nrow in nrows:
data = get_data(nrow, dt)
np.random.shuffle(data)
for dpr in duplicate_rate:
data_c = data.copy()
dupl_val = data_c[0]
num_duplicates = int(dpr * nrow)
dupl_indices = np.random.choice(np.arange(nrow), num_duplicates, replace=False)
data_c[dupl_indices] = dupl_val
for impl in use_range_part:
print(f"{round((i / total_its) * 100, 2)}%")
i += 1
cfg.RangePartitioning.put(impl)
sr = pd.Series(data_c)
execute(sr)
t1 = timer()
# returns a list, so no need for materialization
sr.unique()
tm = timer() - t1
print(nrow, dpr, dt, impl, tm)
result.loc[nrow, (dt, dpr, impl)] = tm
result.to_excel("unique.xlsx")
Measurements with 16 cores being allocated for Modin (MODIN_CPUS=16):
Measurements with 44 cores being allocated for Modin (MODIN_CPUS=4):
Performance measurements for ``.drop_duplicates()``
The performance was measured on randomly generated data using Intel(R) Xeon(R) Gold 6238R CPU @ 2.20GHz (56 cores).
The duplicate rate shows the procentage of duplicated rows in the dataset. The subset size shows the number of
columns being specified as a subset parameter for df.drop_duplicates(). You can learn more about this
micro-benchmark by reading its source code:
Micro-benchmark's source code
import modin.pandas as pd
import numpy as np
import modin.config as cfg
from modin.utils import execute
from timeit import default_timer as timer
import pandas
cfg.CpuCount.put(16)
pd.DataFrame(np.arange(cfg.NPartitions.get() * cfg.MinPartitionSize.get())).to_numpy()
nrows = [1_000_000, 5_000_000, 10_000_000, 25_000_000]
duplicate_rate = [0, 0.1, 0.5, 0.95]
subset = [["col0"], ["col1", "col2", "col3", "col4"], None]
ncols = 15
use_range_part = [True, False]
columns = pandas.MultiIndex.from_product(
[
[len(sbs) if sbs is not None else ncols for sbs in subset],
duplicate_rate,
use_range_part
],
names=["subset size", "duplicate rate", "use range-part"]
)
result = pandas.DataFrame(index=nrows, columns=columns)
i = 0
total_its = len(nrows) * len(duplicate_rate) * len(subset) * len(use_range_part)
for sbs in subset:
for nrow in nrows:
data = {f"col{i}": np.arange(nrow) for i in range(ncols)}
pandas_df = pandas.DataFrame(data)
for dpr in duplicate_rate:
pandas_df_c = pandas_df.copy()
dupl_val = pandas_df_c.iloc[0]
num_duplicates = int(dpr * nrow)
dupl_indices = np.random.choice(np.arange(nrow), num_duplicates, replace=False)
pandas_df_c.iloc[dupl_indices] = dupl_val
for impl in use_range_part:
print(f"{round((i / total_its) * 100, 2)}%")
i += 1
cfg.RangePartitioning.put(impl)
md_df = pd.DataFrame(pandas_df_c)
execute(md_df)
t1 = timer()
res = md_df.drop_duplicates(subset=sbs)
execute(res)
tm = timer() - t1
sbs_s = len(sbs) if sbs is not None else ncols
print("len()", res.shape, nrow, dpr, sbs_s, impl, tm)
result.loc[nrow, (sbs_s, dpr, impl)] = tm
result.to_excel("drop_dupl.xlsx")
Measurements with 16 cores being allocated for Modin (MODIN_CPUS=16):
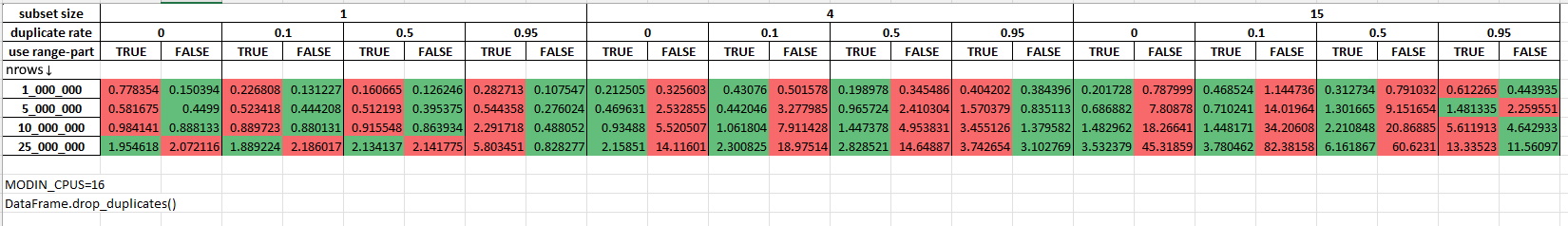
Measurements with 44 cores being allocated for Modin (MODIN_CPUS=44):
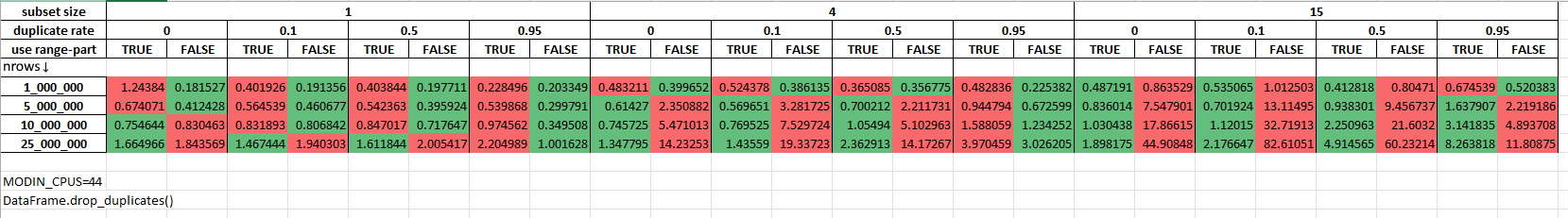
‘.nunique()’#
Note
Range-partitioning approach is implemented only for pd.Series.nunique() and 1-column dataframes.
For multi-column dataframes .nunique() can only use full-axis reduce implementation.
Range-partitioning implementation of ‘.nunique()’’ works best when the input data size is big (more than 5_000_000 rows) and when the output size is also expected to be big (no more than 80% values are duplicates).
Under the spoiler you can find performance comparisons in different scenarios:
Performance measurements for ``.nunique()``
The performance was measured on randomly generated data using Intel(R) Xeon(R) Gold 6238R CPU @ 2.20GHz (56 cores). The duplicate rate shows the procentage of duplicated rows in the dataset. You can learn more about this micro-benchmark by reading its source code:
Micro-benchmark's source code
import modin.pandas as pd
import numpy as np
import modin.config as cfg
from modin.utils import execute
from timeit import default_timer as timer
import pandas
cfg.CpuCount.put(16)
def get_data(nrows, dtype):
if dtype == int:
return np.arange(nrows)
elif dtype == float:
return np.arange(nrows).astype(float)
elif dtype == str:
return np.array([f"value{i}" for i in range(nrows)])
else:
raise NotImplementedError(dtype)
pd.DataFrame(np.arange(cfg.NPartitions.get() * cfg.MinPartitionSize.get())).to_numpy()
nrows = [1_000_000, 5_000_000, 10_000_000, 25_000_000, 50_000_000, 100_000_000]
duplicate_rate = [0, 0.1, 0.5, 0.95]
dtypes = [int, str]
use_range_part = [True, False]
columns = pandas.MultiIndex.from_product([dtypes, duplicate_rate, use_range_part], names=["dtype", "duplicate rate", "use range-part"])
result = pandas.DataFrame(index=nrows, columns=columns)
i = 0
total_its = len(nrows) * len(duplicate_rate) * len(dtypes) * len(use_range_part)
for dt in dtypes:
for nrow in nrows:
data = get_data(nrow, dt)
np.random.shuffle(data)
for dpr in duplicate_rate:
data_c = data.copy()
dupl_val = data_c[0]
num_duplicates = int(dpr * nrow)
dupl_indices = np.random.choice(np.arange(nrow), num_duplicates, replace=False)
data_c[dupl_indices] = dupl_val
for impl in use_range_part:
print(f"{round((i / total_its) * 100, 2)}%")
i += 1
cfg.RangePartitioning.put(impl)
sr = pd.Series(data_c)
execute(sr)
t1 = timer()
# returns a scalar, so no need for materialization
res = sr.nunique()
tm = timer() - t1
print(nrow, dpr, dt, impl, tm)
result.loc[nrow, (dt, dpr, impl)] = tm
result.to_excel("nunique.xlsx")
Measurements with 16 cores being allocated for Modin (MODIN_CPUS=16):
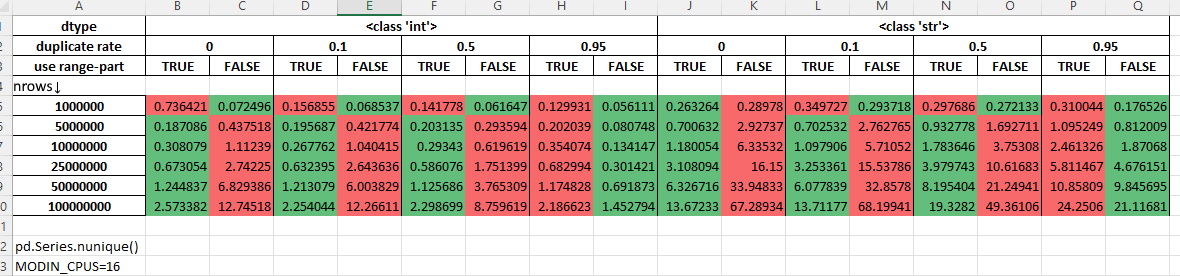
Resample#
Note
Range-partitioning approach doesn’t support transform-like functions (like .interpolate(), .ffill(), .bfill(), …)
It is recommended to use range-partitioning for resampling if you’re dealing with a dataframe that has more than 5_000_000 rows and the expected output is also expected to be big (more than 500_000 rows).
Under the spoiler you can find performance comparisons in different scenarios:
Performance measurements for ``.resample()``
The script below measures performance of df.resample(rule).sum() using Intel(R) Xeon(R) Gold 6238R CPU @ 2.20GHz (56 cores).
You can learn more about this micro-benchmark by reading its source code:
Micro-benchmark's source code
import pandas
import numpy as np
import modin.pandas as pd
import modin.config as cfg
from timeit import default_timer as timer
from modin.utils import execute
cfg.CpuCount.put(16)
nrows = [1_000_000, 5_000_000, 10_000_000]
ncols = [5, 33]
rules = [
"500ms", # doubles nrows
"30s", # decreases nrows in 30 times
"5min", # decreases nrows in 300
]
use_rparts = [True, False]
cols = pandas.MultiIndex.from_product([rules, ncols, use_rparts], names=["rule", "ncols", "USE RANGE PART"])
rres = pandas.DataFrame(index=nrows, columns=cols)
total_nits = len(nrows) * len(ncols) * len(rules) * len(use_rparts)
i = 0
for nrow in nrows:
for ncol in ncols:
index = pandas.date_range("31/12/2000", periods=nrow, freq="s")
data = {f"col{i}": np.arange(nrow) for i in range(ncol)}
pd_df = pandas.DataFrame(data, index=index)
for rule in rules:
for rparts in use_rparts:
print(f"{round((i / total_nits) * 100, 2)}%")
i += 1
cfg.RangePartitioning.put(rparts)
df = pd.DataFrame(data, index=index)
execute(df)
t1 = timer()
res = df.resample(rule).sum()
execute(res)
ts = timer() - t1
print(nrow, ncol, rule, rparts, ts)
rres.loc[nrow, (rule, ncol, rparts)] = ts
rres.to_excel("resample.xlsx")
Measurements with 16 cores being allocated for Modin (MODIN_CPUS=16):
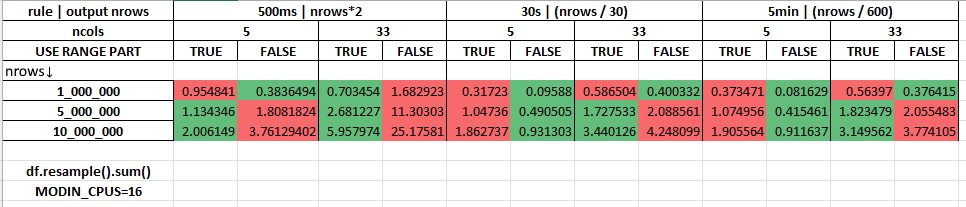
pivot_table#
Range-partitioning implementation is automatically applied for df.pivot_table
whenever possible, users can’t control this.
sort_values#
Range-partitioning implementation is automatically applied for df.sort_values
whenever possible, users can’t control this.
